Caracterización molecular del germoplasma de ñame colombiano utilizando "Selective Amplificaron of Microsatellite Polymorphic Loci" (SAMPL) en condiciones radioactivas
Molecular Characterization of Colombian Yam Germoplasm by "Selective Amplificaron of M i c rósate l lite Polymorphic Loci" (SAMPL)
|
|
||
|
Rev. Colomb. Biotecnol. Vol. VIII N° 2 Diciembre 2006 60-66
|
||
|
|
||
|
Molecular Characterization of Colombian Yam Germoplasm by "Selective Amplificaron of M i c rósate l lite Polymorphic
Loci" (SAMPL)
Caracterización molecular del germoplasma de ñame
colombiano utilizando "Selective Amplificaron of Microsatellite Polymorphic Loci" (SAMPL) en condiciones
radioactivas
Silvia Bustamante R.'", Gustavo Buitrago H'
ABSTRACT
In Colombia, yam (Dioscoreu spp.) has been a basic and traditional crop for small-and medium-scale farmers. It has been an important source of family employment in the Colombian Atlantic Coast for the past several centuries. However, yam is the most fragüe crop regarding market issues and disease susceptibility (e.g. to anthracnose and viruses). At present, the crop is under progressive recovery from a previous drastic reduction in área and production, and apparently allows high profits. Is imperative for the future of the crop, that the rural farmers are supplied with selected varieties, thereby increasing their sustainability and economic yield. However, such varieties are not available, but have to be bred. Breeding of yam in turn is difficult, and presupposes knowledge about the genetic diversity in the breeding germplasm, which is not known. For these reasons, we characterized the Colombian yam varieties used in the fields as well as wild relatives with a molecular marker technique: SAMPL (Selective Amplification of Microsatellite Polymorphic Loci). Positions of 160 unequivocally scorable bands (of which 56 were monomorphic, 35% of total, with an average of 2.8 polymorphic bands per assay) were transformed into a binary character matrix and analyzed. The resulting similarity matrix was transformed into a dendrogram with the UPGMA algorithm. The diversity of the collections with this technique was 0.0471 with a mean similarity of 0.9529.
Key Words: Dioscorea spp, microsatellites, DNA
RESUMEN
En Colombia el æame (Dioscorea spp.) ha sido un cultivo bÆsico tradicional a pequeæa y mediana escala de cultivadores de la Costa AtlÆntica para abastecer el consumo propio y el mercado local, convirtiØndose en una gran fuente de empleo durante muchos aæos, sin embargo el æame es uno de los cultivos mÆs susceptibles a enfermedades (vg. la antracnosis y virus). En la actualidad el cultivo ha ido recuperÆndose progresivamente de una drástica reducción en área, como en producción, permitiendo aparentes resultados benéficos para los productores. Es imprescindible para el futuro del cultivo, que los productores cuenten con variedades seleccionadas para incrementar la sostenibilidad y rentabilidad del cultivo. Sin embargo esas variedades no estÆn disponibles en la naturaleza, sino que tienen que ser mejoradas. El mejoramiento del ñame es difícil y presupone un conocimiento de la diversidad genØtica del germoplasma de Dioscorea en Colombia, la cual no se conoce todavía. Por estas razones nosotros caracterizamos las variedades de ñame colombiano utilizadas en campo, así como las silvestres con una técnica de biología molecular: Selective Amplification Microsatellite Polimorphic Loci (SAMPL). Se detectaron 160 bandas identificables, (de las cuales 56 fueron monomórficas, 35% del total, con un rango de 2.8 bandas polimórficas por análisis). Con la información anterior se construyó una matriz binaria que fue transformada en un dendograma utilizando el algoritmo UPGMA. La diversidad de la colección determinada con esta tØcnica fue de 0.0471 con una media de similaridad de 0.9529.
Palabras clave: Dioscorea spp, microsatélites, DNA
Recibido: octubre 06 de 2006 Aceptado: noviembre 14 de 2006
|
||
|
|
||
|
* Instituto de Biotecnología. Universidad Nacional de Colombia. Bogotá - Colombia.
** Universidad de Frankfurt - Biozentrum Pflanzliche Molekularbiologie Marie-Curie-Str. 9/N200 D- 60439 Frankfurt amMain.
|
||
|
|
||
|
60
|
||
|
|
||
|
|
|||
|
MOLECULAR CHARACTERIZATION OF COLOMBIAN YAM GERMOPLASM
|
|||
|
|
|||
|
INTRODUCTION
Yam (Dioscorea spp) belongs to the family Dioscoreaceae, order Dioscoreales. It is monocotyledonous and constitutes an important staple food crop for millions of people in the humid and subhumid tropics. Tubers provide an important food resource for people from these regions. In Colombia, yam (Dioscorea spp.) has been a basic and traditional crop for small- and medium-scale farmers from the Atlantic Coast (Córdoba, Sucre, Cesar, Atlántico and Bolívar departments). Formerly itwas a self-supporting crop for local markets only, but more recently became an export product. In 1989, pro-duction has been drastically affected by a massive infection of Colletotríchum gloeosporioides, the causative agent of the foliar disease anthracnose (Álvarez, 1991). In subsequent years, smallholders dramatically reduced the acreage for yams, replacing it with other crops such as cassava and plantains. Knowledge of germplasm diversity and relationships among élite materials has a significant impact on the improvement of crop plants generally (e.g. Witcombe, et ál., 2001), and of yam in particular for example the works done in Asian and Africans Dioscorea bulbifera, Guinea yam Dioscorea rotundata-D.cayenensis(Ramser, etál., 1996,1997 respectively) and Jamaican yam germplams (Asemota, et ál., 1996 ), were, using different molecular techniques was possible detected genomic variation for phylogeny and taxonomy studies as well as obtained information about de genome of these collection. Efficient breeding of yam in Colombia can only be achieved if its genetic diversity is known and promising varieties can be selected. This, however, is not the case. For these reasons, we characterized a comprehensive collection of the presently grown Colombian yam varieties with a molecular marker techniques: selective amplification of microsatellite polymorphic loci (SAMPL; Witsenboer, et ál. 1997).
MATERIALS AND METHODS Plant material
Different yam cultivars were collected from fields at different locations at the Caribbean Colombian Coast (Durangoand Padilla, 1998). The samples from África were generously donated by the Internatio-nal Institute of Tropical Agriculture (UTA Ibadan,
|
Nigeria). Dioscorea sansibarensis and D. vittata were provided by in the yam collection of Frankfurt University (table 1).
DNA isolation
DNA was ¡soled according to the procedure of Asemota (1991) with some modifications. For example, dithiotreithol (DDT) was employed for the reduction of quiñones instead of p-mercaptoethanol.
SAMPL analysis
The SAMPL technique makes use of compound microsatellite primers to reveal genetic polymorphisms (Morgante and Vogel, 1994). It combines the high multiplex ratio typical for amplification fragment length polymorphisms (AFLPs; Vos et ál. 1995) with the high level of variability typical for SSRs. The protocol was based on the procedures reported by Morgante and Vogel (1994) and Witsenboer et ál. (1997). Sequences of primers and adapters are given in Table 2. All PCR steps were performed in a Perkin Elmer 9700 thermocycler. Initial restriction enzyme digestions were performed in final volumes of 25 uL containing 200 ng of DNA témplate, 10 units of EcoRI (MBI Fermentas), 33 mM Tris-acetate (pH 7.9), 10 mM magnesium acétate, 66 mM potassium acétate, and 0.1 mg/ mL BSA. After three hours at 37 °C, 10 uL of a ligation mix containing 5.5 ng of EcoRI adapter, 1 mM ATP, 33 mM Tris-acetate (pH 7.9), 10 mM magnesium acétate, 66 mM potassium acétate, 0.1 mg/mL BSA and 2 U T4 DNA ligase (MBI Fermentas) were added. After overnight incubation at 37 °C, ligation-restriction reactions were diluted 1:4 with bi-distilled sterile water. Pre-amplifications were performed in 20 uL reactions containing 5 uL of diluted ligation products, 75 mM Tris-HCl (pH 9), 20 mM (NH4)2SO4, 0.01 % Tween-20, 1.5 mM MgCI2, 200 uM of dNTPs, 0.5 uM EcoRI adapter-specific primer, and 0.5 U Taq polymerase (Biotherm, Gene Craft). The PCR program consisted of an initial denaturation step at 94 °C (120 s), 34 cycles at 94 °C (30 s), 50 °C (30 s), and 72 °C (60 s). After pre-amplification, 3 uL aliquots were separated in 1% agarose gels and stained with ethidium bromide. The remaining samples were diluted 1: 9 and stored at -20 °C until use. Compound microsatellite primers (see table 2) were end-labelled with [y^P]-
|
||
|
|
|||
|
61
|
|||
|
|
|||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Rev. Colomb. Biotecnol. Vol. VIII N° 2 Diciembre 2006 60-66
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Table 1. Origin of plant material
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
* c.v : cultivar number or ñame of clone
Note: UTA, International Institute for Tropical Agricultura, Ibadan, Nigeria
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
62
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
MOLECULAR CHARACTERIZATION OF COLOMBIAN YAM GERMOPLASM
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
ATP and polynucleotide kinase (MBI Fermentas) using standard procedures (Sambrook, et ál. 2000). Selective amplification was performed in 10 uL reactions containing 2 uL of diluted pre-amplified fragments, 0.5 uM labelled microsatellite primer, 5 uM EcoRI adapter-specific primer, 400 uM of dNTPs, 75 mM Tris-HCl (pH 9), 20 mM (NH4)2SO4, 0.01% Tween-20, 1.5 mM MgCl2, and 0.5 U Taq polymerase (Biotherm, Gene Craft). The PCR program consisted of an initial denaturation step at 94°C(120s), 11 cyclesat94°C(30s),65°C(30 s) [with a touch-down of -0.7 °C each cycle], and 72 °C (60 s), followed by 25 cycles at 94 °C (30 s), 56 °C (30 s), and 72 °C (60 s). Final extensión was at 72 °C (120 s). Products were separated on 4% sequencing gels and autoradiographed (Sambrook, et ál., 2000). The compound microsatellite primers used for the selective amplification were designed and provided by Dr. Peter Winter (unpublished data). Additionally, a newly designed adapter-primer ("Eco RI short") was employed in combination with TA-rich selective primers (Palacios, et ál. 2002).
|
Data analysis
Unequivocally scorable SAMPL bands were transformed into a binary character matrix, with 1 for the presence, and 0 for the absence of a band at a particular position. Pairwise distances were directly computed with the STATISTICA sofware package (1984-1999 Stat Soft. Inc) using the percent disagreement. The final dendrogram was constructed using UPGMA cluster algorithm (Dhillon and Ishiki, 1999).
RESULTS AND DISCUSSIONS
Genomic DNAs from 36 Colombian and 9 African D. alata accessions, 12 Colombian and 11 African D. rotundata accessions, one accession each of D. trífida, D. bu l bífera, D. vittata and D. sansibarensis, respectively, were analyzed by SAMPL with a set of 5 compound microsatellite primers and a modified EcoRI adapter primer (Palacios, et ál., 2002) (table 2). Distinct polymorphic
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Table 2. Primers sequences
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
63
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||
|
Rev. Colomb. Biotecnol. Vol. VIII N° 2 Diciembre 2006 60-66
|
|||
|
|
|||
 |
|||
|
|
|||
|
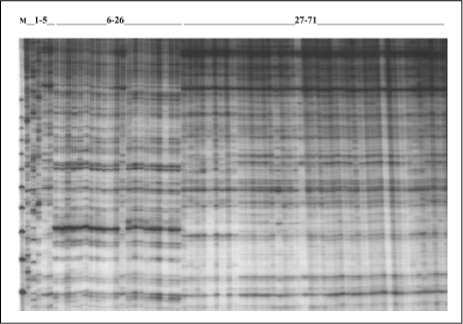
Figure 1. SAMPL analysis of the whole D. alata and D. rotundata collections using SAMPL primer38. M: Molecular weigh marker 25pb, MBI Fermentas, 1- 5: D. trífida, D. bulbifera, D. vittata, D. sansibarensis. Car1, 6 - 26: D. rotundata, 26 - 71: D. alata.
|
|||
|
|
|||
|
banding patterns were obtained with all primers, as exemplified by a subset of samples and SAMPL primer 38 (figure. 1). Positions of 160 unequivocally scorable bands (of which 56 were monomorphic, 35% of total, with an average of 2.8 polymorphic bands per assay) were transformed into a binary character matrix(1 forpresence, Oforabsence)andanalyzed. The resulting similarity matrix was transformed into a dendrogram with the UPGMA algorithm. The diversity of the collections with this technique was 0.0471 with a mean similarity of 0.9529.
The obtained SAMPL banding patterns clearly reflect the lack of genetic diversity observed within and between sampling sites. Basically the clusters observed obey to species separation. African accessions are found in cióse similarity with
|
Colombian individuáis, sincethewayof propagation applied by the farmers is in principle clonal, some accessions are still identical with the African "very likely" ancesters (Coursey and Martin, 1970; Coursey, 1976a, 1976b). On the other hand, somaclonal variation, which is the only source of variability, shows also to be rather low. Within the same species, form hundred-sixty scored characters, only four polymorphic bands were observed among individuáis form separated clusters. The same type of results were observed with DAF markers (Bustamante, et ál., 2003) were was evident the low genetic variability. Is important to develop varieties carrying as many different genes for resistance as possible, in order to provide stable resistance against a broad spectrum of the pathogens. Therefore, is imperative to extend the
|
||
|
|
|||
|
64
|
|||
|
|
|||
|
|
|||
|
MOLECULAR CHARACTERIZATION OF COLOMBIAN YAM GERMOPLASM
|
|||
|
|
|||
|
yam germplasms in Colombia by plant breeding or introducing new varieties.
ACKNOWLEDGEMENTS
Research of the authors was supported by grants from GTZ (grant 95.2072.7-001.00) and BMZ (89.7860.3-01.130). S. B acknowledges a fellowship of UNESCO (Paris, France), Stiftung zur Fórderung der internationaler wissenschaftlicher Bezierungen (Frankfurt am Main, Germany), and PBA Fundation and Córdoba University for Colombian yam Collection. The authors also acknowledge Diego Riaño (Instituto de Biotecnología, Universidad Nacional de Colombia BogotÆ, Colombia) and Carlos Molina (Plant Molecular Biology, Biocentre, University of Frankfurt, Frankfurt am Main / Germany) for their contributions.
BIBLIOGRAFÍA
Álvarez, A. 1991. Control de la antracnosis en æame. Instituto Colombiano Agropecuario (ICA). El Carmen de Bolívar. Documento de trabajo. Pág. 15.
Asemota, H.N. 1991. A fast, simple, and efficient mini-scale method for the preparation of DNA from tissues of yam (Dioscorea spp.). Plant Mol Biol Rep. 13: 214-218.
Asemota, H.N.; Ramser, J.; Lopéz-Peralta, C; Weising, K.; Kahl, G. (1996). Genetic variation and cultivar identification of Jamaican yam germplasm by random amplified polymorphic DNA analysis. Euphytica. 92: 341-351.
Bustamante, S.; Guzmán, M.; Buitrago, G. 2001. Molecular characterization of some species and varieties of yam present on the Colombian Atlantic Coast. Rev Col Bio. 2: 38-43.
Bustamante, S.; Guzmán, M.; Buitrago, G. 2003. Molecular characterization of Colombian yam germplasm by DNA amplification fingerprinting DAR Rev Col Bio. 5: 57-63.
Coursey D.G; Martin, R.W. 1970. The past and future of yams as crop plants. In: Proceedings Second Symposium of Tropical Root Crops, Hawai i. 1: 87-90; 99-101.
Coursey, D.G. 1976a. Yams Dioscorea spp. (Dioscoraceae). In: Evolution of Crop Plants. N.W. Simmonds, ed. Longmans, London an New York. Pp. 70-74.
Coursey, D.G. 1976b. The origins and domestication of yams in África. In: Origins of African Plant
|
Domestication. J.R. Harían; J.M.J. de Wet and A.B.L. Stemler, eds. Mouton, The Hague. pp. 383-403.
Durango, R.; Padilla A. 1998. Caracterización morfológica de un banco de germoplasma de ñame Dioscorea spp recolectado en la costa norte colombiana. Trabajo de grado Universidad de Córdoba, Montería, Colombia.
Dhillon, N.P.S.; Ishiki, K. 1999. Genomic variation and genetic relationship in Ipomoea spp. Plant Breed. 118: 161-165.
Morgante, M.; Vogel, J. 1994. Compound microsatellite primers for the detection of genetic polymorphisms. US Patent Application. 08/326456.
Nei, M.; Li, W.H. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 76: 5269-5273.
Paglia, G; Morgante, M. 1998. PCR-based multiplex DNA fingerprinting techniques for the analysis of conifer genomes. Mol Breed. 4: 173-177.
Palacios, G.; Bustamante, S.; Molina, C.; Winter, P; Kahl, G. 2002. Electrophoretic identification of new genomic profiles with a modified SAMPL technique based on AT/AAT polymorphic repeats. Electrophoresis. 23: 3341-3345.
Ramser, J.; López-Peralta, C; Wetzel, R.; Weising, K.; Kahl, G. 1996. Genomic variation and relationships in aerial yam (Dioscorea bulbifera L) detected by ramdom amplified polymorphic DNA. Genome. 39: 17-25.
Ramser, J.; Weising, K.; Lopez-Peralta, C; Terhalle, W.; Terauchi, R.; Kahl, G. 1997. Molecular marker based taxonomy and phylogeny of Guinea yam (Dioscorea rotundata - D. cayenensis). Genome. 40: 903-915.
Ramser, J.; Weising, K.; Chikaleke, V.; Kahl, G. 1997. Increased informativeness of RAPD analysis by detection of microsatellite motifs. BioTechniques. 23: 285-290.
Sambrook, J.; Fritsch, E.F.; Maniatis, T. 2000. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Vivek, B.S.; Simón, PW. 1999. Linkage relationships among molecular markers and storage root traits of carrot (Daucus carota L ssp. sativus). Theor Appl Genet. 99: 58-64.
|
||
|
|
|||
|
65
|
|||
|
|
|||
|
|
||
|
Rev. Colomb. Biotecnol. Vol. VIII N° 2 Diciembre 2006 60-66
|
||
|
|
||
|
Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Van production systems in Nepal and India.
de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Euphytica. 122: 575-588.
Peleman.J.; Kuiper, M.¡ Zabeau, M. 1995. Witsenboer, H.; Vogel, J.; Michelmore, R.W. 1997.
AFLP: a new technique for DNA fingerprinting. identificaron, genetic localization, and allelic
Nucí Acids Res. 21: 4407-4414. d¡vers¡ty of selectively amp|¡f¡ed polymorphic loci
Witcombe, J.R.; Joshi, K.D.; Rana, R.B.; Virk, D.S. ¡n lettuce and wNd relatives (Lactuca spp.).
2001. Increasing genetic diversity by Genome. 40: 923-936.
participatory varietal selection in high potential
|
||
|
|
||
|
66
|
||
|
|
||
Cómo citar
APA
ACM
ACS
ABNT
Chicago
Harvard
IEEE
MLA
Turabian
Vancouver
Descargar cita
Visitas a la página del resumen del artículo
Descargas
Licencia
Derechos de autor 2006 Revista Colombiana de Biotecnología

Esta obra está bajo una licencia internacional Creative Commons Atribución 4.0.
Esta es una revista de acceso abierto distribuida bajo los términos de la Licencia Creative Commons Atribución 4.0 Internacional (CC BY). Se permite el uso, distribución o reproducción en otros medios, siempre que se citen el autor(es) original y la revista, de conformidad con la práctica académica aceptada. El uso, distribución o reproducción está permitido desde que cumpla con estos términos.
Todo artículo sometido a la Revista debe estar acompañado de la carta de originalidad. DESCARGAR AQUI (español) (inglés).


















